This page is an appendix to our paper:
- Édouard Thiel and Rita Zrour
[ paper pdf 300k ]
Exact Medial Axis based on Flake Digital Circles.
In DGMM 2024, Discrete Geometry and Mathematical Morphology, Florence, april 2024, rejected.
Demonstration programs
The programs are based on the OpenCV library, available for Linux, windows and MacOS systems, and the small GUI helper tool Astico2D.
| FlakesMA-v0.4.tgz | Download the source code in C++ language, licence CC-BY, and some examples of images. |
| README.txt | Documentation for installation and compilation. |
Two programs are provided. The first comes with a keyboard-driven GUI:
$ ./flakes-ma-gui
Usage : ./flakes-ma-gui [-mag w h] [-thr threshold] [-o image_res] image_src [other options]For instance, by running ./flakes-ma-gui IMAGES/seal_K3.pgm we get:
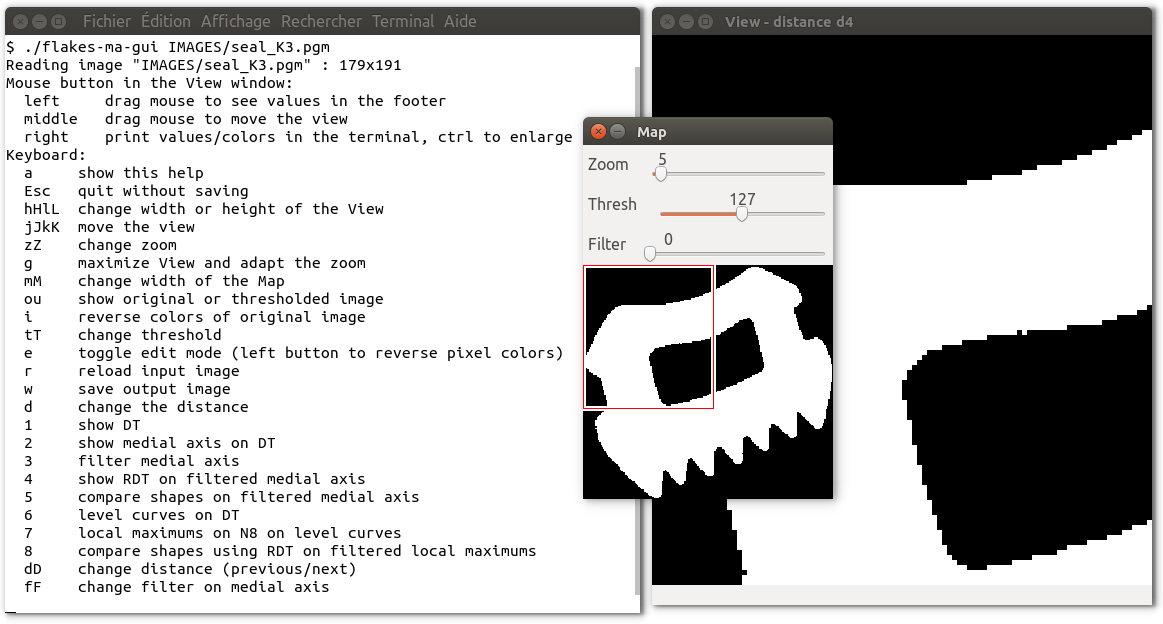
Press the sequence: g 2 D D D D D D D to see the medial axis
for different distances.
The available distances are: d4, d8,
the weighted distances d2,3, d3,4, d5,7,11,
the squared Euclidean distance dE²,
and the proposed flake distances d'F1 and d'F0 :
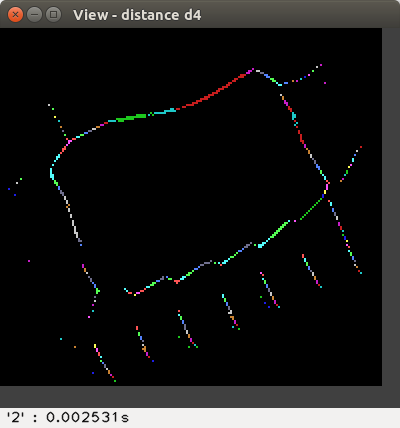
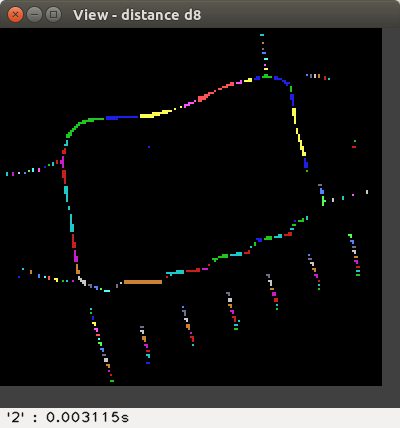
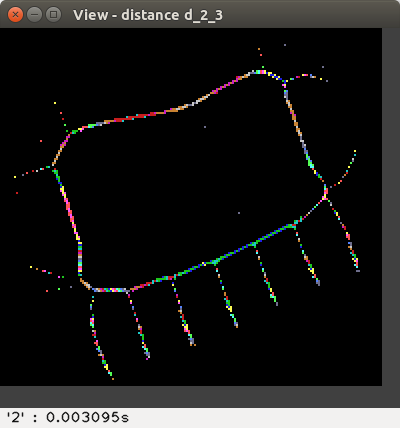
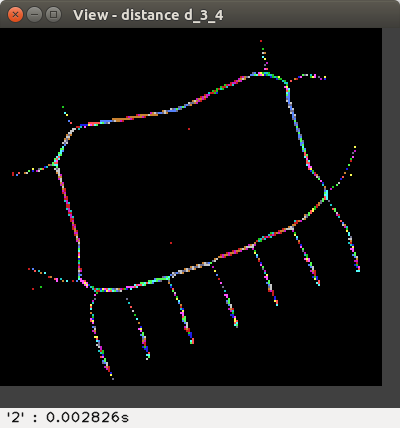
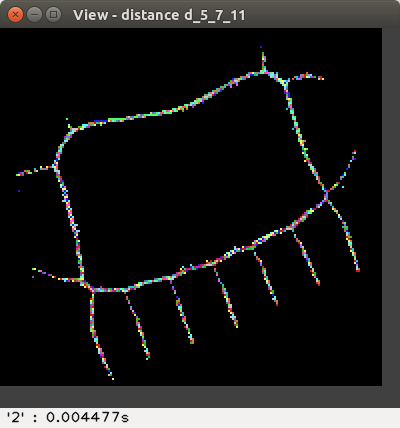
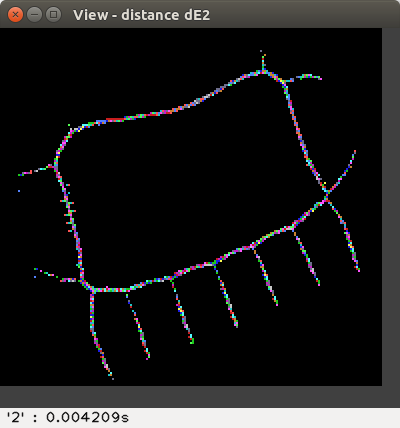
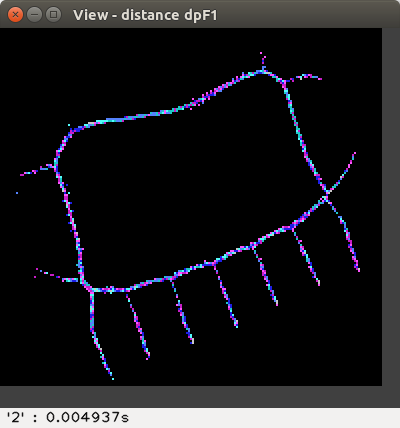
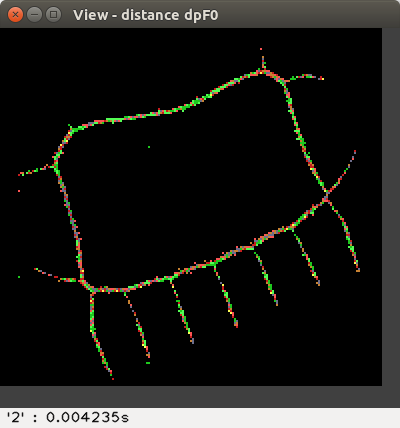
The distance values are represented with false colours
(VGA: 0 black, 1 blue, 2 green, 3 cyan, 4 red, 5 magenta, 6 brown,
7 light gray, 8 dark gray, 9 light blue, 10 light green, 11 light cyan,
12 light red, 13 light magenta, 14 yellow, 15 blue, ..., 255 white, ...).
Left-click to see the distance value at the bottom-right of the View window,
or right-click to print a neighbourhood in the terminal:
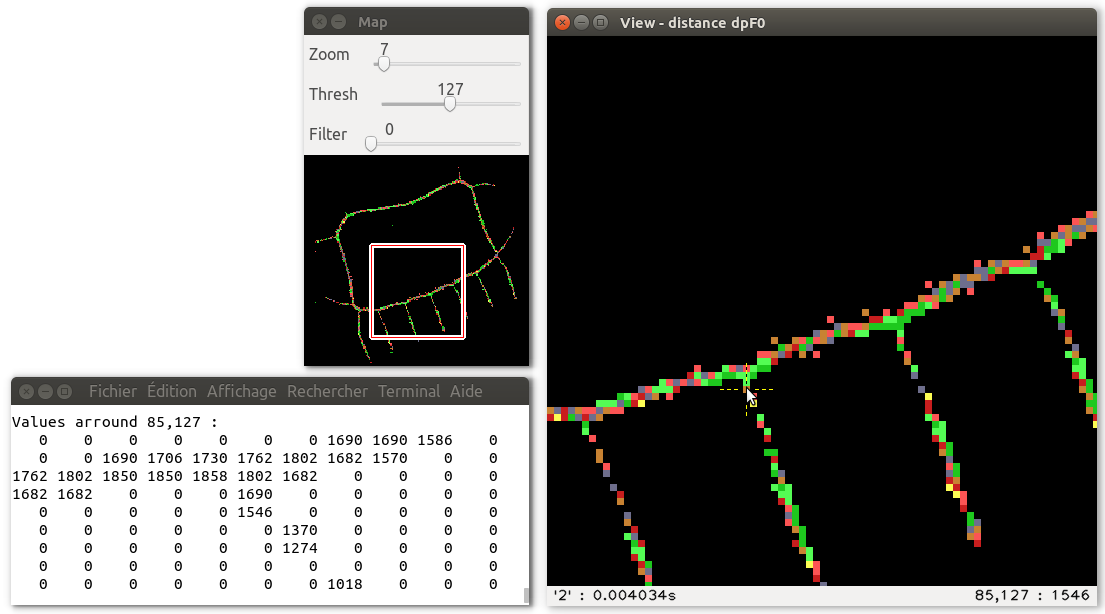
The transformation 5 shows the medial axis other the shape,
and allows to test the reversibility of MA after a reverse DT;
the background is labelled 0 (black),
the medial axis points are at 11 (light cyan),
the recovered shape pixels after RDT are at 8 (dark gray),
missing shape pixels are at 12 (light red),
and extra pixels are at 14 (yellow).
The MA is then fully reversible if they are neither light red nor yellow pixels:
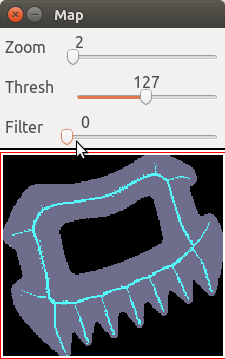


Using the slider "Filter" or the keys f F, we can set a
threshold to filter the medial axis, and see the non-recovered pixels.
For instance with a threshold of 300 we obtain:
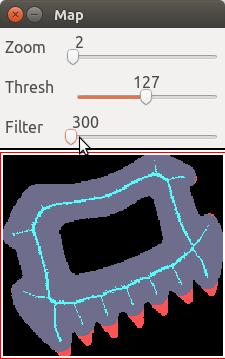

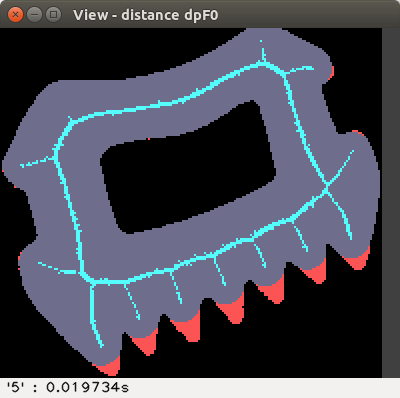
The image can be modified: type e, then left click or drag inside
the shape; type r to reload, or e to leave edit mode.
The transformations 6 7 8 are experimental:
6 displays the level curves,
7 computes an MA on the level curve using d8,
8 acts like 5 to measure reversibility.
The result is a quasi-MA, that is more or less a subset of the exact MA,
but lacks reversibility on the border of the shape.
The program flakes-ma-gui accepts gray levels and colour images
as an input; colour images are converted in gray levels, then gray levels
are thresholded (by default at 127) to get a binary image.
Run ./flakes-ma-gui IMAGES/cat1.jpg, type i u
(or run any transform, e.g. 5),
then modify the threshold using t T or the slider "Thresh".
The second program is a command-line tool:
$ ./flakes-ma-cli
Usage: ./flakes-ma-cli [-d dist_name] actions
dist_name:
'd4', 'd8', 'd_2_3', 'd_3_4', 'd_5_7_11', 'dE2', 'dpF1', 'dpF0'
actions:
[-show-dt] -comp-mlut L
[-i in1] [-o out2] [-size h w] [-add|-sub] [-p x y v]... -comp-rdtTo compute the LUT mask for the distance d'F1 for L=100, type:
$ ./flakes-ma-cli -d dpF1 -comp-mlut 100
Distance: dpF1
Computing MLutg...
GreatestRadius for L = 100 is 38808
compute_lut_mask for L = 100, Rtarget = 38808
i= 1 (x,y)= ( 1, 0) added for R= 1 visible
i= 2 (x,y)= ( 1, 1) added for R= 5 visible
i= 3 (x,y)= ( 2, 1) added for R= 369 visible
i= 4 (x,y)= ( 3, 1) added for R= 1233 visible
i= 5 (x,y)= ( 3, 2) added for R= 1413 visible
i= 6 (x,y)= ( 4, 1) added for R= 4765 visible
i= 7 (x,y)= ( 5, 1) added for R= 5337 visible
i= 8 (x,y)= ( 5, 2) added for R= 9601 visible
i= 9 (x,y)= ( 4, 3) added for R= 10229 visible
i= 10 (x,y)= ( 5, 4) added for R= 14965 visible
i= 11 (x,y)= ( 5, 3) added for R= 16417 visible
i= 12 (x,y)= ( 7, 1) added for R= 20457 visible
i= 13 (x,y)= ( 6, 1) added for R= 24085 visible
i= 14 (x,y)= ( 7, 2) added for R= 26821 visibleThe Mlut points up to L = 1000 are presented in this file, for the distances dE², d'F1 and d'F0. These data complete the results shown in the Figure 8 in the paper. All detected points are visible points.
The option -comp-rdt allows to create images containing reverse balls.
For instance, to create a 100x150 image containing reverse balls for the distance
d'F0, of centre (50,50) and radius 1600, and of center (90,60) and radius 2500,
type:
$ ./flakes-ma-cli -d dpF0 -o tmp1.png -size 100 150 -p 50 50 1600 -p 90 60 2500 -comp-rdt
Distance: dpF0
Creating image 100x150 ...
Drawing pixels:
50 50 1600
90 60 2500
Computing RDT ...
Making binary result ...
Saving image tmp1.png ...
successfully saved.
$ ./flakes-ma-gui tmp1.png

Here is how to generate an image containing a reverse ball of radius 46 pixels for each of the 8 available distances:
$ ./flakes-ma-cli -d d4 -size 100 800 -o tmp2.png -p 50 50 46 -comp-rdt ;
./flakes-ma-cli -d d8 -i tmp2.png -o tmp2.png -add -p 150 50 46 -comp-rdt ;
./flakes-ma-cli -d d_2_3 -i tmp2.png -o tmp2.png -add -p 250 50 92 -comp-rdt ;
./flakes-ma-cli -d d_3_4 -i tmp2.png -o tmp2.png -add -p 350 50 138 -comp-rdt ;
./flakes-ma-cli -d d_5_7_11 -i tmp2.png -o tmp2.png -add -p 450 50 230 -comp-rdt ;
./flakes-ma-cli -d dE2 -i tmp2.png -o tmp2.png -add -p 550 50 2116 -comp-rdt ;
./flakes-ma-cli -d dpF1 -i tmp2.png -o tmp2.png -add -p 650 50 8464 -comp-rdt ;
./flakes-ma-cli -d dpF0 -i tmp2.png -o tmp2.png -add -p 750 50 8464 -comp-rdt ;
./flakes-ma-gui tmp2.png

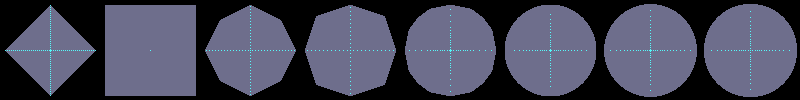
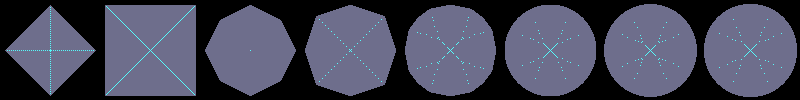
Using flakes-ma-gui, we can then observe that the MA is the
sole centre of the ball for the corresponding distance, and contains
other points when the ball does not belong to the balls distance set:
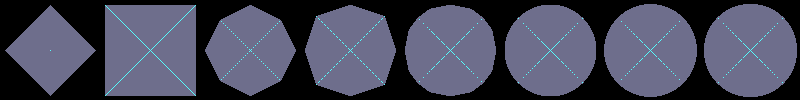 d4
d4
 d8
d8
 d2,3
d2,3
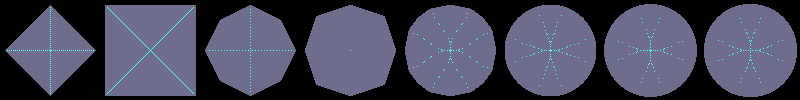 d3,4
d3,4
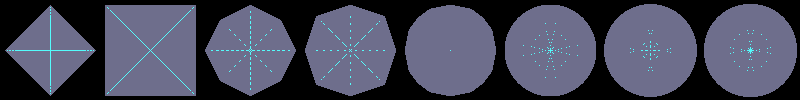 d5,7,11
d5,7,11
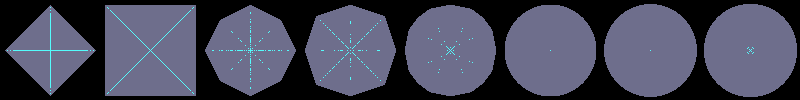 dE²
dE²
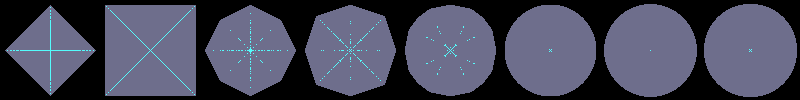 d'F1
d'F1
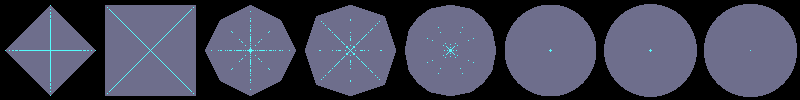 d'F0
d'F0